Docker TerraForm and SQL
1/18/26
Difference b/w an Data Engineer and and AI Engineer
- Data Engineer -> making sure all the data is captured, ready for analysis , clean and can be used.
- AI Engineer -> work on the AI aspect of the product , do prompt tuning, validate your model, verifiable rewards etc.
Docker For Data Engineering:
- Docker is a way to separate what we have in our host machine and the application (that requires other software dependencies).
- What we have in Docker container is completely isolated from host machine.
- What we run has no effect from the host machine and has no effect from the host machine.
- Simplest Docker Command to check if Docker installed correctly:
docker run hello-world- Can run with interactive terminal as
docker run -it ubuntu- Whatever we do here is isolated from the host machine
- To come back we do
docker run -it ubuntu - But when we do this , the problem is the command is then not found.
- Everytime we create an docker container from an Docker image.
- An instance of the Docker image is created -> it contains a complete snapshot of the operating system.
- It is stateless and doesn’t preserve the state.
- Instead of Ubuntu we can also use the direct python image of 3.13 directly also.
- When installing the Python 3.13 image :
- python -> image name
- 3.13.11-slim -> is the tag name
- If we wanted the entry point to be a bash session and not python repl directly:
docker run -it --entrypoint=bash python:3.13.11-slim
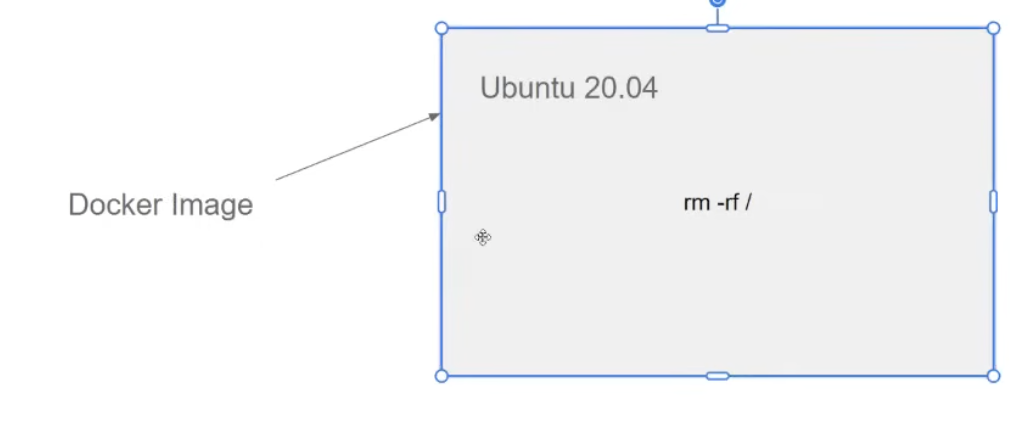
- We can see stopped containers (which are saved somewhere as)
docker ps -a- even if we do
rm -rf / - lists all the docker images that we executed and they have all the state
- can remove all the files as
docker ps -aq-> gets the id- then run as `
docker rmdocker ps -aq
- then run as `
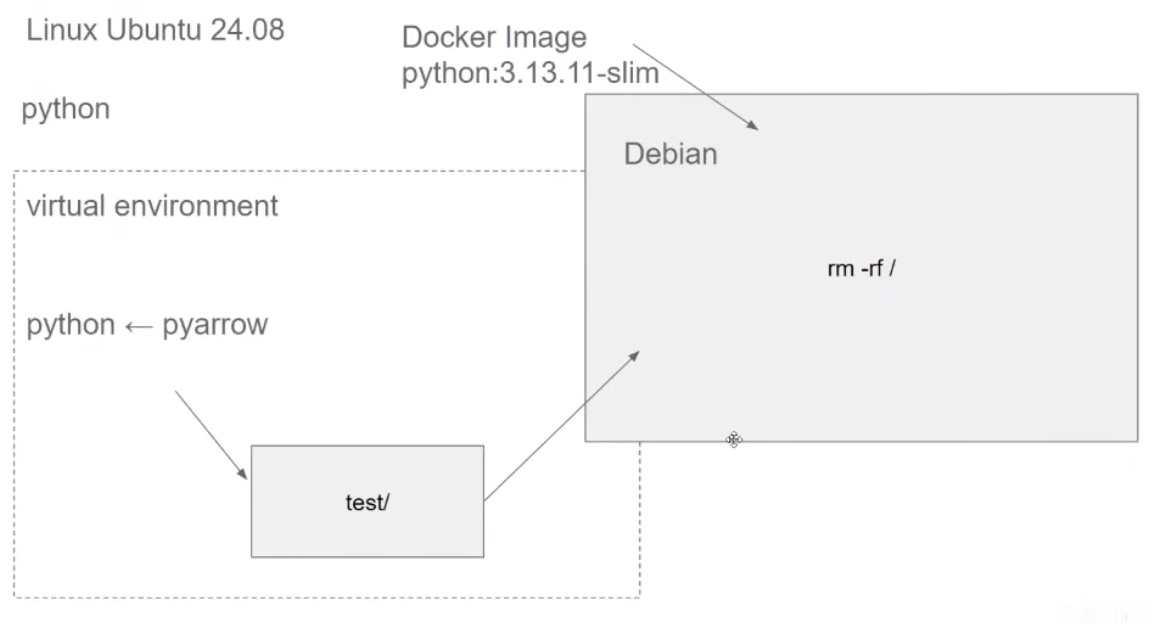
- How to preserve state ?
- let us have a dir
testand it contains files in that directory - what if we need to have access to the files from our docker container ?
- let us have a dir
- Executing a file from inside volumes:
- We have a folder
test/that should be available for both host machine and the container. - Using volume mapping as :
docker run -it --entrypoint=bash -v $(pwd)/test:/app/test python:3.13.11-slim - where
$(pwd)/test/is the local fs directory where it is kept - and
/app/testis where it is being mapped to in Docker. - remember this should be executed from
$(pwd)and not inside from$(pwd)/test
- We have a folder
Data Pipelines:
- A data pipeline is a service that receives data as input and outputs more data.
- Eg: reading a CSV file, transforming the data somehow and storing it as a table in a PostgreSQL database.
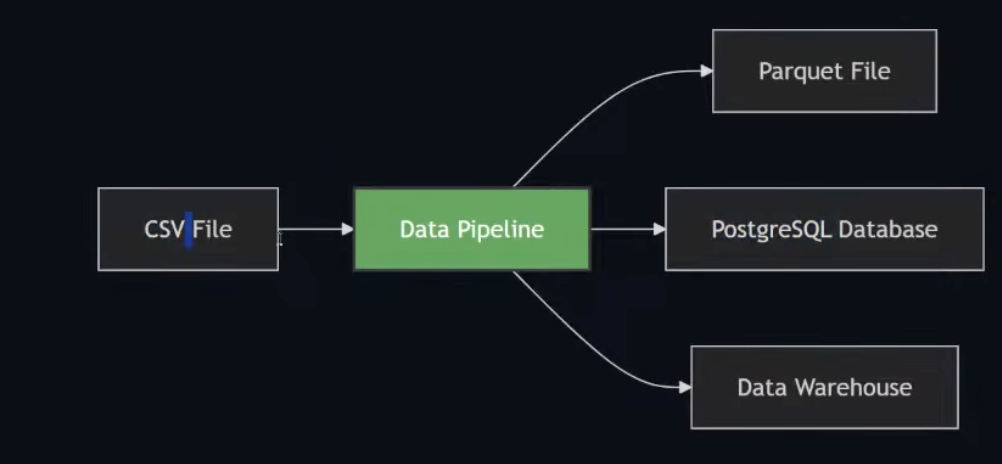
- We have CSV files and parses the CSV files and creates the parquert files -> takes in some input and produces some output.
- Anything that can take data from one and put in some destination is called a data pipeline.
Building a basic pipeline in pipeline/pipeline.py:
- arguments to check data in month 12 using the sys module
- using pandas to then visualize the data in the given month
- then saving the data to parquet -> parquet is a binary format for data. optimized for csv
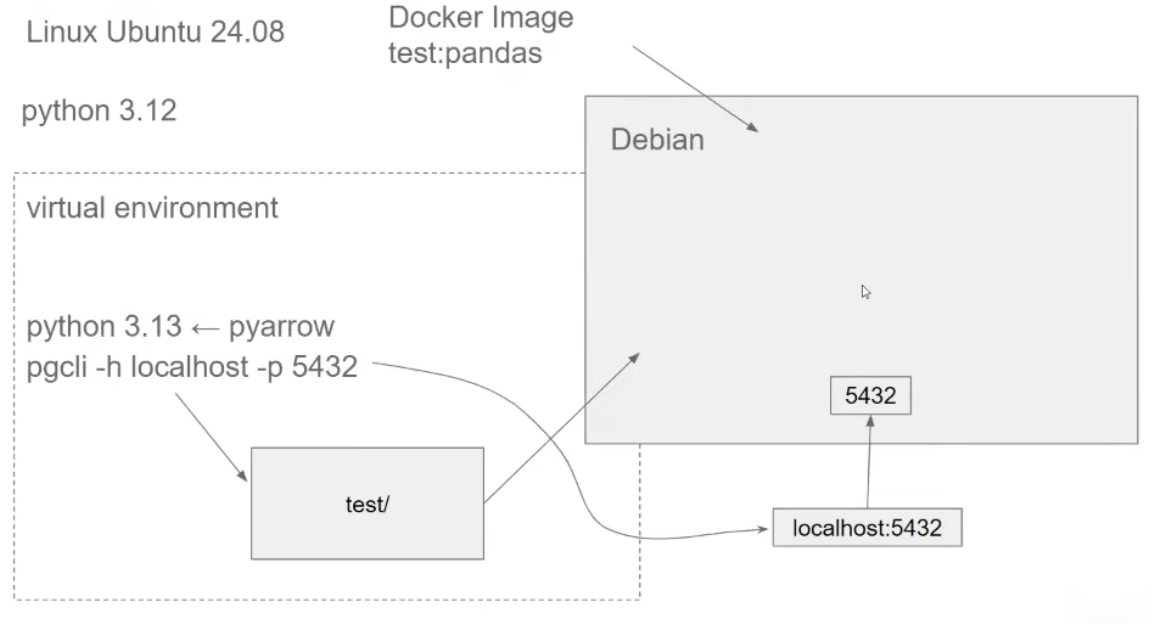
- want a isolated environment for setting up pyarrow and parquet.
- create an virtual environment for the environment , different from the virtual environment we have for the docker image.
- easier to isolate one project from another
- using uv to manage creating virtual environments
uv init --python 3.13- it creates its own python and it will use the uv’s python to manage the project and to add the dependencies.
- now we create an new docker image , python 3.13 , pyarrow, pandas and when we run docker , we run this pipeline.py
Dockerfileis a special file that tells how we are gonna create the Docker container.- Run as `docker build -t test:pandas .
-tis the tag name that we need to pass.is the directory we are targetting ; where Dockerfile is specifically- Then can run as
docker run -it --entrypoint=bash --rm test:pandas- when we finish the docker sessions, we wont have the state saved somewhere
- no have any dangling things left
- Using
ENTRYPOINTwe can redefine where we want it to run it from next time.docker run -it --rm test:pandas 12
- in our local machine we used
uvbut in our docker image we didnt.- update our docker image to copy from the host system to the docker image
COPY --from=ghcr.io/astral-sh/uv:latest /uv /bin/- after doing this uv becomes available.
- after copying this we run
RUN uv sync --lockedto have the same dependencies on the docker container - so the python env in our local and in docker will exactly will be the same.
- we can pass the uv run command in the entrypoint
- or better to use it in the env path
ENV PATH="/code/.venv/bin:$PATH"
Adding Postgres to our flow:
- Docker can help with specific version of Postgres for our setup.
- Setting up basic postgres on docker:
docker run -it --rm -e POSTGRES_USER="root" -e POSTGRES_PASSWORD="root" -e POSTGRES_DB="my_taxi" -v ny_taxi_postgres_data:/var/lib/postgresql -p 5432:5432 postgres:18-eflag are used to setup env variables- for postgres we are configuring it with a user , password and database name.
- this is different from every container , the potential variables can be used.
ny_taxi_postgres_datawill be a inner volume for Docker that we will not be able to see- with it we can preserve our data
- we can put whatever data that we want , and save it and next time it would be there
- The concept is the same with volume mapping
- we have a host machine and we map it to a container
- With port mapping
- we have a port in host machine and we map it to a port in the container.
- we sent a request to
localhost:5432, is actually sent to whatever is running to inside in the container.
- pgcli
- python tool to intertact with postgres
- adding it as a dev dependency to the uv project.
- not a main dependency, using this dependency to just interaction with dev,not dependent on production use case.
uv run pgcli -h localhost -p 5432 -u root -d ny_taxi
- Flow of things till now :
- Even if we disconnect from it now, it will still have all the data till now.
Interactive Analysis of Our Data using Jupyter
- adding
jupyteras a dev dependency to see and analyze the data into a jupyter notebook and to see the dependency. - we want to use pandas to read a csv file — which is present in remote — nyt taxi dataset , which is in parquet.
- useful engineering tip -> added the prefix to set the main route to download from and url to point to specific dataset we want
- then use
pd.read_csv()to download the specific url - can then look at the data using
df.head() - now our goal is to put this data into postgres.
- parquet also schemas in the datatype, but csv doesnt
- so pandas is trying to infer this type somehow
- so we use a custom datatype , so to remove the type not matching warning
dtype = { "VendorID": "Int64", "passenger_count":"Int64", "trip_distance":"float64", "RatecodeID":"Int64", "store_and_fwd_flag":"string", "PULocationID":"Int64", "DOLocationID":"Int64", "payment_type":"Int64", "fare_amount":"float64", "extra":"float64", "mta_tax":"float64", "tip_amount":"float64", "tolls_amount":"float64", "improvement_surcharge":"float64", "total_amount":"float64", "congestion_surcharge":"float64" }- to add this in our postgres , we would need an ORM :
sqlalchemyand also thepsyciog2-binarywhich is the postgres driver for python. - first we create an engine in our jupyter notebook to ingest the data.
from sqlalchemy import create_engine engine = create_engine("postgresql://root:root@localhost:5432/ny_taxi")- add the schema only (without inserting data)
df.head(0).to_sql(name='yellow_taxi_data',con=engine,if_exists='replace')
- then use
- now to save this data into postgres , we should save them in chunks , otherwise the datasets could be huge and we dont want to load them up in memory.
- we define an iterator :
df_iter = pd.read_csv(url,dtype=dtype,parse_dates=parse_dates, iterator=True,chunksize=100000)- and then using a for loop we can iterate and push the data into the database.
- can then insert and check progress using
tqdmfor df_chunk in tqdm(df_iter): df_chunk.to_sql(name='yellow_taxi_data',con=engine,if_exists='append')
Optimized Data Ingestion
- generate a script from the jupyter notebook and create an ingestion pipeline
- refactored into clean ingestion pipeline that can be run with a single command.
python pipeline/ingest_data.py
Making a Better command line interface using click
- Using
clickto make a better command line interface to use our parameters to get the data and make it better.
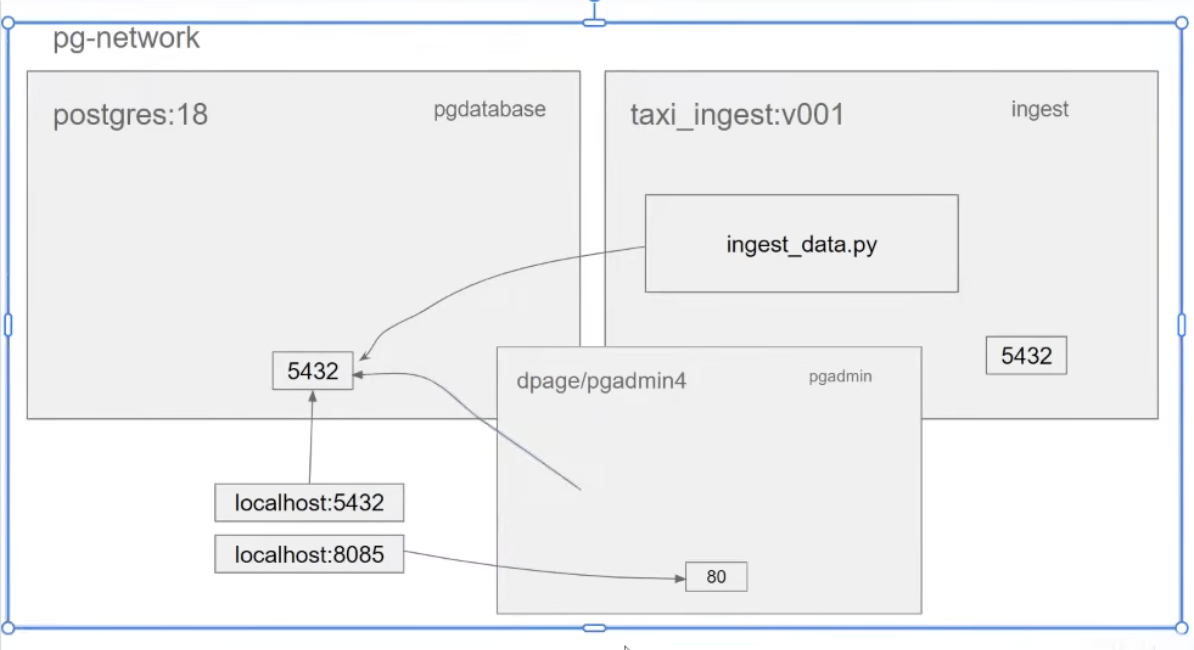
Architecture till now:
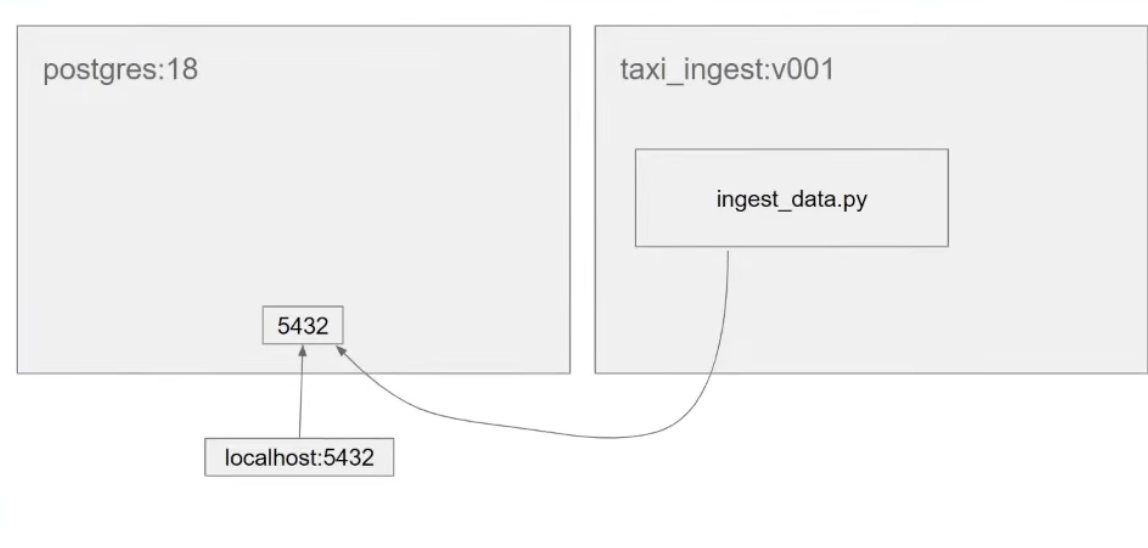
- when we try to connect to the localhost , it is trying to connect to the localhost internally to docker not to our local.
- need to create using
docker network - things within the same network can see each other.
pgdatabaseinpostgres:18containertaxi_ingest:v001iningestcontainer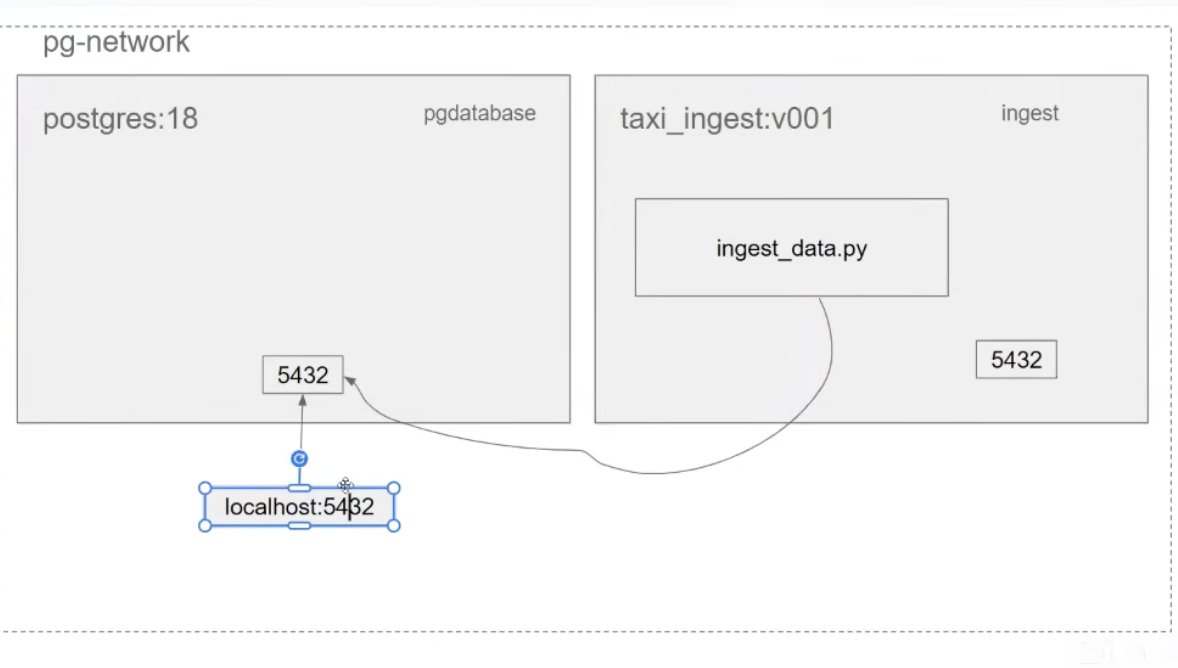
- they can then see each other and can also access ports of each other.
- after resolving this, we added pgAdmin to run in the same network to get a better UI.
- After adding pgadmin:
docker-compose
- to execute both the services db up and setup pgAdmin together.
- creates volumes and networks